
 |
|||||
| Introduction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cluster-Buster is our third generation program for finding clusters of pre-specified motifs in nucleotide sequences. The main application is detection of sequences that regulate gene transcription, such as enhancers and silencers, but other types of biological regulation may be mediated by motif clusters too. Cluster-Buster may be used via our web server, or downloaded for use on your local computer. We also provide a downloadable program Cluster-Trainer for estimating optimal motif weights and gap parameters for Cluster-Buster. On Dec 4, 2007 we fixed a bug on `dust' option. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Publication | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Martin C Frith, Michael C Li, and Zhiping Weng (2003). Nucleic Acids Research, 31(13):3666-8. (Abstract) Here is some supporting data for our publication, which demonstrates that Cluster-Buster overcomes a fundamental problem of hidden Markov model algorithms. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample Output | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
This example shows output of Cluster-Buster applied to GenBank sequence AY007685, which contains the human TERT gene encoding the catalytic subunit of telomerase. The first diagram shows an overview of motif cluster locations in the sequence, along with protein-coding regions (CDS) annotated in the GenBank record: Homo sapiens telomerase catalytic subunit (TERT) and sodium channel-like protein genes, complete cds.   KEY: motif cluster protein-coding Next, detailed information for each cluster is printed. Here are the details for the second strongest cluster, corresponding to the second tallest green bar in the overview: Cluster: 21602 to 22627 Score: 25 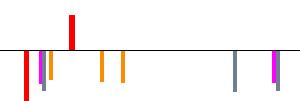
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FAQ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Q: What do the scores mean? A: The scores are log likelihood ratios. The cluster score is log [ prob(cluster sequence given that it's a cluster of real sites) / prob(cluster sequence given that it's random DNA) ]. The motif score is log [ prob(motif sequence given that it's a real site) / prob(motif sequence given that it's random DNA) ]. The higher the better. Q: How high is high enough? A: Unfortunately there's no easy answer. You could try running Cluster-Buster on some control sequences (matched for GC content, etc.) and seeing what scores you get. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Credits | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Programming and documentation: Martin C Frith Website authoring and design: Michael C Li |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Comments and questions to Martin Frith | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||