1601 bp
1601 bp
1600 bp
1601 bp
1600 bp
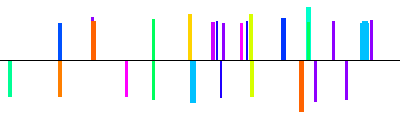
1601 bp
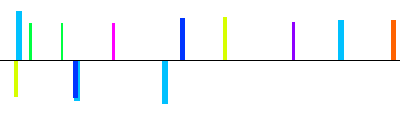
1601 bp
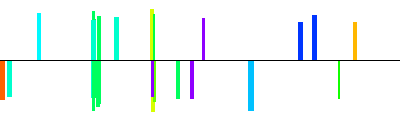
| Motif | Overall Raw Score | P(userbgseq) |
|---|---|---|
| SRF | -5.01 | 0 |
| RXR-VDR | -2.06 | 0 |
| Brachyury | -2.94 | 0 |
| SU | -1.09 | 0 |
| GATA-1 | -0.0993 | 0 |
| GATA-3 | -0.821 | 0 |
| Yin-Yang | 0.568 | 0.001 |
| Max | -0.94 | 0.001 |
| Myc-Max | -1.77 | 0.001 |
| TBP | -2.12 | 0.002 |
| GATA-2 | -0.819 | 0.004 |
| Tal1beta-E47S | -0.0676 | 0.005 |
| CF2-II | -1.36 | 0.006 |
| Bsap | 1.1 | 0.007 |
| Ubx | -1.32 | 0.993 |
| SQUA | -1.02 | 0.996 |
| E4BP4 | -4.17 | 0.999 |
| Broad-complex | -0.645 | 1 |
| Broad-complex | 0.292 | 1 |
| S8 | -1.52 | 1 |
| Irf-1 | -2.79 | 1 |
| AP2alpha | 0.214 | 1 |
| NM_022427|gpr88|1600bp 1601 bp | 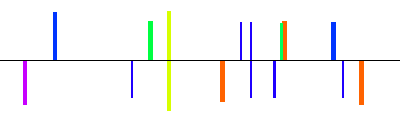 | + |
| - | ||
| NM_078477|KLF16|1600bp 1601 bp | 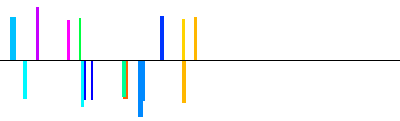 | + |
| - | ||
| NW_000028|A2AR|m1A|1600bp 1600 bp | 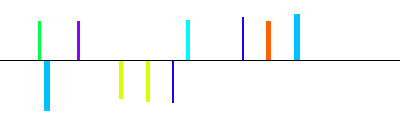 | + |
| - | ||
| NM_008313|htr4|1600bp 1601 bp |  | + |
| - | ||
| NM_011268|rgs9|1600bp 1600 bp | 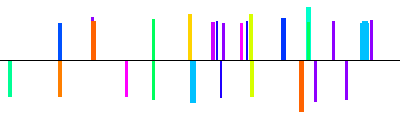 | + |
| - | ||
| NM_013643|ptpn5|1600bp 1601 bp | 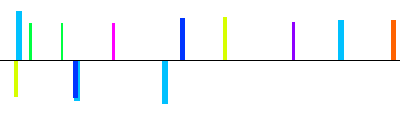 | + |
| - | ||
| NT_039474|ARPP-19|ProSeq_37|contig|1600bp 1601 bp | 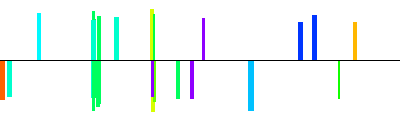 | + |
| - |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| actttagttta | 96 - 106 | - | Broad-complex | 7.44 | 0.001147 |
| acataaaaagccag | 215 - 228 | + | SQUA | 7.85 | 0.009753 |
| taatt | 527 - 531 | - | S8 | 6.28 | 0.02654 |
| accatc | 594 - 599 | + | Yin-Yang | 6.4 | 0.004551 |
| accatc | 605 - 610 | + | Yin-Yang | 6.4 | 0.004551 |
| ccgacagctggt | 671 - 682 | + | Tal1beta-E47S | 8.06 | 0.008427 |
| gacagctggtgg | 673 - 684 | - | Tal1beta-E47S | 8.37 | 0.00668 |
| actatggtactcacac | 884 - 899 | - | SU | 6.86 | 0.01125 |
| aatta | 964 - 968 | + | S8 | 6.25 | 0.02712 |
| taatt | 1003 - 1007 | - | S8 | 6.28 | 0.02654 |
| aatta | 1004 - 1008 | + | S8 | 6.25 | 0.02712 |
| taatt | 1097 - 1101 | - | S8 | 6.28 | 0.02654 |
| gtatatttat | 1122 - 1131 | + | CF2-II | 6.11 | 0.02501 |
| ctctgagaaaagaagt | 1132 - 1147 | + | SU | 6.33 | 0.01743 |
| ctgaaaaaggtatt | 1328 - 1341 | + | SQUA | 6.32 | 0.02639 |
| taatt | 1372 - 1376 | - | S8 | 6.28 | 0.02654 |
| agtgggtttgccacat | 1439 - 1454 | - | SU | 7.47 | 0.007143 |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| agagcactggaacctgacca | 43 - 62 | + | Bsap | 7.12 | 0.003605 |
| atgacacccag | 96 - 106 | - | Brachyury | 6.42 | 0.02898 |
| taaacatgatg | 146 - 156 | + | Broad-complex | 8.72 | 0.0001807 |
| tcctactt | 272 - 279 | + | Broad-complex | 6.53 | 0.001764 |
| accatc | 318 - 323 | + | Yin-Yang | 6.83 | 0.001428 |
| ttcacaacttg | 326 - 336 | - | Brachyury | 7.69 | 0.01178 |
| atta | 341 - 344 | - | Ubx | 6.54 | 0.0007044 |
| atta | 369 - 372 | - | Ubx | 6.54 | 0.0007044 |
| ctgttcactttt | 492 - 503 | - | Irf-1 | 6.1 | 0.03128 |
| ttcacttttgccatac | 495 - 510 | - | SU | 6.36 | 0.017 |
| cgaactctacaaact | 557 - 571 | - | RXR-VDR | 9.44 | 0.001047 |
| caaactcgatcagct | 566 - 580 | - | RXR-VDR | 6.7 | 0.01194 |
| ccaagaatggcatt | 642 - 655 | + | SQUA | 7.23 | 0.01546 |
| gcgcacgtggc | 730 - 740 | + | Myc-Max | 6.81 | 0.01271 |
| gcacgtggcc | 732 - 741 | - | Max | 7.07 | 0.007574 |
| ggccacgtgc | 779 - 788 | + | Max | 7.07 | 0.007574 |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| gccatc | 156 - 161 | + | Yin-Yang | 6.41 | 0.004359 |
| tgtttatgctccactgtgtt | 177 - 196 | - | Bsap | 8.42 | 0.0008972 |
| gccctaagg | 310 - 318 | + | AP2alpha | 6.38 | 0.01069 |
| gccagatggacc | 479 - 490 | - | Tal1beta-E47S | 6.42 | 0.02651 |
| aacagctcgttg | 588 - 599 | - | Tal1beta-E47S | 6.85 | 0.01964 |
| taatt | 691 - 695 | - | S8 | 7.05 | 0.01017 |
| gtaggtgacaa | 748 - 758 | + | Brachyury | 6.54 | 0.02571 |
| aatta | 971 - 975 | + | S8 | 7.1 | 0.009035 |
| tcgtgggatcctttgg | 1067 - 1082 | + | SU | 6.36 | 0.017 |
| ggggccctgctgcggagaga | 1179 - 1198 | + | Bsap | 7.6 | 0.002147 |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| gccatc | 72 - 77 | + | Yin-Yang | 6.5 | 0.00339 |
| aatactatataaatta | 246 - 261 | + | TBP | 7.7 | 0.01108 |
| aatta | 257 - 261 | + | S8 | 6.31 | 0.02605 |
| acaaatttaataat | 326 - 339 | + | SQUA | 6.23 | 0.0278 |
| taatt | 336 - 340 | - | S8 | 6.25 | 0.02712 |
| aatta | 337 - 341 | + | S8 | 6.31 | 0.02605 |
| taatt | 351 - 355 | - | S8 | 6.25 | 0.02712 |
| gctttcactgtg | 363 - 374 | - | Irf-1 | 7.35 | 0.01328 |
| gccatc | 400 - 405 | + | Yin-Yang | 6.5 | 0.00339 |
| agatag | 430 - 435 | + | GATA-3 | 6.06 | 0.004919 |
| aaaactaagca | 690 - 700 | + | Broad-complex | 6.32 | 0.006287 |
| agcagatggagt | 1106 - 1117 | - | Tal1beta-E47S | 6.71 | 0.02166 |
| agtcacgtgg | 1253 - 1262 | + | Max | 8.58 | 0.002197 |
| agtcacgtggt | 1253 - 1263 | + | Myc-Max | 8.56 | 0.002529 |
| gtcacgtggtc | 1254 - 1264 | - | Myc-Max | 8.62 | 0.00244 |
| tcacgtggtc | 1255 - 1264 | - | Max | 9.98 | 0.0002184 |
| cacaggtggtca | 1270 - 1281 | - | Tal1beta-E47S | 6.02 | 0.03518 |
| agatag | 1306 - 1311 | + | GATA-3 | 6.06 | 0.004919 |
| gatggc | 1312 - 1317 | - | Yin-Yang | 6.47 | 0.003704 |
| acaactgaaagc | 1364 - 1375 | + | Irf-1 | 6.59 | 0.02267 |
| gcccagagg | 1497 - 1505 | + | AP2alpha | 6.3 | 0.01206 |
| gcccctggc | 1513 - 1521 | - | AP2alpha | 6.93 | 0.00463 |
| gcccctggc | 1513 - 1521 | + | AP2alpha | 6.77 | 0.005866 |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| attttcgatttt | 35 - 46 | - | Irf-1 | 6.02 | 0.03341 |
| agctcacataa | 234 - 244 | - | E4BP4 | 6.06 | 0.01894 |
| gctcacataagg | 235 - 246 | + | SRF | 6.12 | 0.03313 |
| gcccagggg | 366 - 374 | + | AP2alpha | 7.03 | 0.003974 |
| ccaggggaaacgagca | 368 - 383 | + | SU | 6.41 | 0.01618 |
| aaatagtg | 504 - 511 | - | Broad-complex | 6.02 | 0.003392 |
| gtatatctat | 610 - 619 | - | CF2-II | 6.59 | 0.01689 |
| gtatatctat | 610 - 619 | + | CF2-II | 6.75 | 0.01462 |
| gagcaggtggc | 756 - 766 | + | Myc-Max | 7.55 | 0.006744 |
| tggccactctggaggcatct | 763 - 782 | - | Bsap | 7 | 0.004254 |
| caaacaaatac | 846 - 856 | + | Broad-complex | 6.26 | 0.00687 |
| aatta | 867 - 871 | + | S8 | 6.44 | 0.02377 |
| taatt | 883 - 887 | - | S8 | 6.23 | 0.02757 |
| gcctaaggg | 889 - 897 | + | AP2alpha | 6.11 | 0.01616 |
| tactagtt | 962 - 969 | + | Broad-complex | 6 | 0.003475 |
| aatta | 987 - 991 | + | S8 | 6.44 | 0.02377 |
| tctacatctggt | 999 - 1010 | + | Tal1beta-E47S | 7.51 | 0.01259 |
| tacatctggtat | 1001 - 1012 | - | Tal1beta-E47S | 6.14 | 0.03251 |
| tcttataaagaaat | 1128 - 1141 | + | SQUA | 6.92 | 0.01876 |
| agctcctttcccactg | 1197 - 1212 | - | SU | 8.59 | 0.002473 |
| ttctgtatataaagag | 1226 - 1241 | + | TBP | 8.74 | 0.005432 |
| gtatataaag | 1230 - 1239 | + | CF2-II | 6.22 | 0.02321 |
| ccttggggc | 1259 - 1267 | - | AP2alpha | 6.91 | 0.004737 |
| gccggacgc | 1329 - 1337 | + | AP2alpha | 6.43 | 0.009898 |
| gcgcgcggc | 1383 - 1391 | - | AP2alpha | 6.6 | 0.007625 |
| ggtgcgagggagagtcgagg | 1442 - 1461 | + | Bsap | 6.06 | 0.01154 |
| ggagagtcgaggggtggagg | 1450 - 1469 | + | Bsap | 6.37 | 0.008564 |
| agtcgaggggtggagggcgg | 1454 - 1473 | + | Bsap | 6.01 | 0.01205 |
| gccctgcgc | 1482 - 1490 | + | AP2alpha | 6.59 | 0.007699 |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| gccatatggctt | 59 - 70 | - | Tal1beta-E47S | 6.13 | 0.03279 |
| ggctttctggagcggagaga | 66 - 85 | + | Bsap | 8.07 | 0.001218 |
| gccatc | 120 - 125 | + | Yin-Yang | 6.11 | 0.008831 |
| gccatc | 247 - 252 | + | Yin-Yang | 6.11 | 0.008831 |
| atttctttctttct | 296 - 309 | - | SQUA | 6.23 | 0.0278 |
| ctttctttcttccttttttt | 300 - 319 | - | Bsap | 6.76 | 0.00556 |
| ttcttttt | 452 - 459 | + | Broad-complex | 6.09 | 0.003093 |
| ctgtcccgctgcataagtca | 651 - 670 | - | Bsap | 7.28 | 0.003148 |
| acagaaaaagaagg | 725 - 738 | + | SQUA | 6.88 | 0.01899 |
| catccatctgtc | 894 - 905 | + | Tal1beta-E47S | 7.03 | 0.01747 |
| gccctaggc | 1171 - 1179 | + | AP2alpha | 6.29 | 0.01221 |
| ggagcacgcaaggagagcgg | 1356 - 1375 | + | Bsap | 6.5 | 0.007426 |
| gtgtgagaagccggga | 1569 - 1584 | + | SU | 6.49 | 0.01532 |
| sequence | position | strand | motif | score | Ranking_ratio |
|---|---|---|---|---|---|
| tgacatgttcacacag | 3 - 18 | - | SU | 6.59 | 0.01412 |
| caggatatatattgtg | 30 - 45 | - | TBP | 6.09 | 0.03195 |
| acaggtgtgaa | 153 - 163 | + | Brachyury | 7.73 | 0.0117 |
| aaaaatatatatacac | 367 - 382 | + | TBP | 6.58 | 0.02344 |
| aaatatatatacacac | 369 - 384 | - | TBP | 6.29 | 0.0283 |
| atatatatac | 371 - 380 | - | CF2-II | 8.35 | 0.002706 |
| atatatatac | 371 - 380 | + | CF2-II | 8.07 | 0.003608 |
| atatatacac | 373 - 382 | - | CF2-II | 6.23 | 0.023 |
| atatatacac | 373 - 382 | + | CF2-II | 6.8 | 0.01411 |
| atacatatat | 389 - 398 | - | CF2-II | 7.76 | 0.005769 |
| acatatatat | 391 - 400 | - | CF2-II | 6.92 | 0.01263 |
| atatatatat | 393 - 402 | - | CF2-II | 7.17 | 0.009828 |
| atatatatat | 393 - 402 | + | CF2-II | 7.17 | 0.009828 |
| atatatataa | 395 - 404 | + | CF2-II | 6.35 | 0.02059 |
| cttgctatataagaac | 461 - 476 | + | TBP | 7.12 | 0.01658 |
| tggccagctggc | 605 - 616 | + | Tal1beta-E47S | 8.42 | 0.006312 |
| gccagctggcca | 607 - 618 | - | Tal1beta-E47S | 8.61 | 0.005485 |
| ccagctggc | 608 - 616 | - | AP2alpha | 6.03 | 0.01835 |
| gccatc | 615 - 620 | + | Yin-Yang | 7.57 | 0.0001967 |
| ccatcc | 616 - 621 | - | GATA-1 | 6.84 | 0.0002271 |
| gaatatatac | 708 - 717 | - | CF2-II | 6.42 | 0.01969 |
| accagcggc | 765 - 773 | - | AP2alpha | 6.4 | 0.01033 |
| gcccccaag | 810 - 818 | + | AP2alpha | 6.9 | 0.004762 |
| tcttcctgcctcagattcct | 995 - 1014 | - | Bsap | 8.46 | 0.0008251 |
| ttaaataaggaaat | 1196 - 1209 | + | SQUA | 6.19 | 0.02823 |
| ctaaatatagacaa | 1252 - 1265 | + | SQUA | 7.4 | 0.014 |
| ctatct | 1354 - 1359 | - | GATA-3 | 6.33 | 0.003036 |
| tatcacatgg | 1416 - 1425 | + | Max | 6.2 | 0.01563 |
Page execution time: 310 seconds.